Brave New Medicine: How an Enzyme Created with Synthetic DNA May Enable the First Treatment for Celiac Disease
By Lisa Howard
The way Justin Siegel describes it, ordering synthetic DNA online is almost as easy as ordering a pair of shoes from Zappos. “You just type it in — or if the protein has been sequenced at one point, we can copy and paste — order it, and it shows up five days later.”
Siegel is an assistant professor of chemistry, biochemistry and molecular medicine at UC Davis. He is also one of the co-founders of PvP Biologics, a university spinoff that is seeking U.S. Food and Drug Administration approval for a therapeutic enzyme for the treatment of celiac disease, an autoimmune disorder triggered by ingesting gluten.
Enzymes that can potentially digest gluten are not a new idea for treating celiac disease. There are food supplements you can buy in health food stores that claim to digest gluten. There have also been credible therapeutic versions, including an oral treatment that combined two enzymes, SC PEP and EP-B2, created in Chaitan Khosla’s lab at Stanford University.
But even though an enzyme might demonstrate an ability to break down gluten in a test tube, the human digestive system is much more complex.
Siegel points out that people don’t just eat gluten; they eat a meal. In addition to a bun, they might also eat a hamburger with cheese and some fries and maybe drink a beer or a big chocolate milkshake. All in one sitting.
It is extremely difficult for an enzyme supplement to be able to zero in on just the gluten mixed up in a complicated slurry of food. And an additional complication is that the gluten needs to be digested in the stomach before it reaches the small intestines where it triggers an immune reaction in people with celiac disease.
“The immunogenic portion of gluten is just a tiny piece of the protein. It’s this little peptide,” explained Siegel. “The needle-in-a-haystack analogy is perfect because this peptide looks just subtly different from all the other proteins present in a meal, but that subtle difference is what triggers a whole immune response for people with celiac disease.”
Undergraduate competition launches new enzyme
During the summer of 2011, Siegel and Ingrid Swanson Pultz, who were both pursuing Ph.D.s at the University of Washington, were advisors for an undergraduate team that would be competing at the International Genetically Engineered Machine (iGEM) competition that fall.
Held every year in Boston, iGEM is essentially the Olympics of synthetic biology. Teams of undergraduates attend from across the world. They are expected to “solve real-world challenges by building genetically engineered biological systems with standard, interchangeable parts.”
The undergraduates had friends who couldn’t eat wheat — meaning, among other things, they had to forgo college rituals like beer and pizza — so the real-world problem the students were interested in solving was gluten intolerance.
The team researched existing literature and found a naturally occurring enzyme called kumamolisin that could survive acidity, such as that found in the stomach, and also target a specific repeating amino acid “motif” for destruction. The iGEM team set out to engineer the enzyme to switch this target motif to the one thought to be the trigger for gluten intolerance.
To construct possible variations of their enzyme, the team used an online multiuser video game called Foldit, which is based on a software program called Rosetta created in the lab of David Baker at the University of Washington.

Justin Siegel. an assistant professor of chemistry, biochemistry and molecular medicine at UC Davis, is also one of the co-founders of PvP Biologics, a university spinoff that is seeking U.S. Food and Drug Administration approval for a therapeutic enzyme for the treatment of celiac disease. (UC Davis/Karin Higgins)
“Foldit is like 3-D Tetris for folding proteins,” explained Siegel. “Fireworks go off, energy changes. Red things show up when something is bad, blue stuff shows up when it’s good.”
Foldit’s intuitive visual interface allows players to rapidly interact and model proteins without needing to learn about complex molecular interactions through years of study. “Users focus on manipulating molecular interactions while gaining an intuition of how these molecular interactions are working,” said Siegel.
Using Foldit, the students created roughly 100 variations of kumamolisin. They inserted synthetic DNA of those variations into a harmless strain of E. coli (some E. coli cause illness but most are harmless) and then tested the resulting proteins made in the microbes for efficacy in breaking down the peptide motif associated with immunogenic gluten. They then took the top performers — variations that were the most effective at breaking down gluten — and combined those traits as well.
The team ended up with an enzyme that was significantly better, in gastric conditions, at breaking down gluten than SC PEP, which at that time had been licensed to Alvine Pharmaceuticals for consideration as an oral therapy for celiac disease.
And the students managed to do all of this in just one summer. Perhaps not surprisingly, the University of Washington team walked away with the grand prize.
Synthetic DNA melds engineering with biology
The enzyme they created, now named KumaMax, would not exist without synthetic biology.
Scientific advances in genomics have increased exponentially in recent years. Genetic sequencing allows researchers to read DNA. Genetic engineering techniques allow researchers to move genes from one organism to another. CRISPR/CAS9 is letting researchers edit DNA within an organism. And with the ability to create and utilize synthetic DNA, a piece of genetic code can be built to specification.
With synthetic DNA, researchers are sometimes trying to replicate a piece of DNA that already exists — something that occurs in nature — but often the DNA is a fabrication, like what the University of Washington students created by making different iterations — mutations — of the original kumamolisin enzyme.
One way to think about synthetic biology is that it’s a discipline that melds engineering (what a piece of DNA can do) with biology (what an organism can produce). Biological cells become almost like little factories that can produce a variety of products. And although synthetic biology has been around for a while, the range of applications for it are growing. Pieces of DNA are being inserted into microbes to do everything from growing collagen that can be used for an artificial leather, to producing spider silk from yeast, to encoding and storing digital information that could one day replace computer hard drives.
Getting DNA wasn’t always this easy. “Before synthetic DNA, you had to go out into nature and clone genes directly from the naturally occurring organism, and then hope the way the organism genetically encoded the protein of interest would be functional in easy-to-use model organisms,” said Siegel. “It had low success rates, and it was very difficult. It would commonly be an entire Ph.D. project to clone one gene and characterize it.”
Working with synthetic DNA is much easier. When it arrives at Siegel’s lab in the Genome and Biomedical Sciences Facility at UC Davis, the synthetic DNA is in a clear test tube, no refrigeration needed. Because he orders it “sequenced verified,” he trusts that the genetic code is exactly what he typed in.
The next step is to get the DNA into an organism. Siegel generally uses E. coli. “We know how to work with it, it grows fast and it’s easy to use.” Siegel explains that there are several ways to get E. coli to take up the external DNA. “We use electrical shocks or we use temperature fluctuations. Then, once the E. coli take up that external DNA we’ve encoded, it starts producing the proteins we are researching.”
And although the technique of growing an artificially created protein in E. coli to create a new therapeutic for celiac disease may seem fantastical, synthetic biology is likely to dramatically change how drugs are created in the future. And KumaMax may end up being the first viable treatment for celiac disease, which can take a heavy toll on the body.
Gluten-free diet is only treatment for celiac disease
Here’s what happens when people with celiac disease eat gluten: Their immune system goes on the attack. Their body treats gluten, a protein found in wheat, barley and rye, like it’s some sort of hostile invader, and the resulting inflammatory response damages the lining of the small intestine.
Sooraj Tejaswi, M.D., an associate clinical professor in UC Davis’ Division of Gastroenterology and Hepatology, uses a carpet analogy to describe the result of this immune system assault.
“Under a microscope, the villi in the small intestines, which help with nutrient absorption, should look like a shag carpet. But in people with celiac disease, the villi get destroyed faster than they can regenerate, so you end up with pretty flat mucosa,” essentially more like some nubby Berber carpet, “that can no longer absorb nutrients,” said Tejaswi.

Sooraj Tejaswi, M.D., an associate clinical professor in UC Davis’ Division of Gastroenterology and Hepatology, treats patients with celiac disease. “It’s a shock for patients when they come to clinic and we tell them ‘you have to have to avoid gluten.’ It’s like you are handing them a harsh sentence for life,” said Tejaswi. (UC Davis/Lisa Howard)
Being able to break down ingested gluten before it has a chance to cause a reaction could be a true breakthrough for the disease. But for the enzyme created at the University of Washington to be a true treatment, it will need to be able to break down the gluten in the stomach before it has a chance to enter the intestines where it causes so much damage.
In addition to intestinal damage, celiac disease is associated with a wide variety of health problems including vitamin and mineral deficiencies (which lead to other health problems, like osteoporosis and anemia) lactose intolerance, infertility, nervous-system disorders, a failure to thrive in children and other complications. In rare cases, celiac disease can lead to small intestinal lymphomas.
The disease is usually first diagnosed with a blood test known as Tissue Transglutaminase Antibodies, which tests positive in about 98 percent of people with celiac disease who are eating gluten. Next is an endoscopy to biopsy the small intestine — to look for those nubby villi — in order to confirm the diagnosis.
Classified as an autoimmune disorder, celiac disease is considered to be vastly underdiagnosed, meaning the majority of people who have it probably don’t know they have it. Studies estimate that the disease affects about 1 percent of the population worldwide.
“It’s somewhat ingrained in the medical community that it’s not a disease of adults or older populations. That it’s more a pediatric disease,” said Tejaswi. “But more and more we realize it can manifest in adulthood. About 25 percent of patients with celiac disease are diagnosed after the age of 60.”
There is a strong genetic component to celiac disease, but the disease is not as simple as cause and effect. “Everyone who has celiac disease has these biomarkers, but even if you have the biomarker you don’t necessarily have celiac disease,” said Siegel. “Or to put it another way, just because you have DQ2 and DQ8 doesn’t mean you have celiac.”
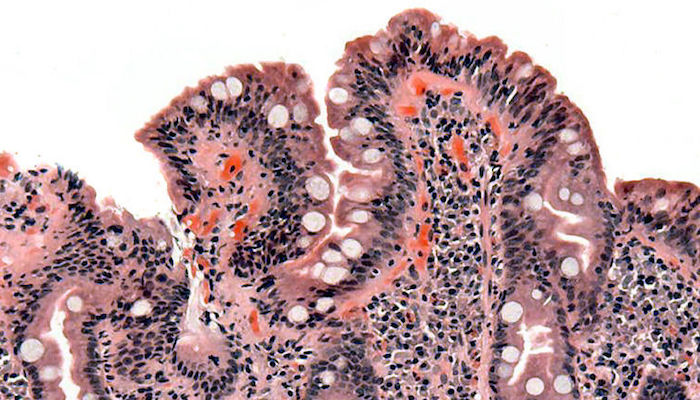
“Berber carpet vs. shag.” For people with celiac disease, an inflammatory reaction in response to gluten causes the shortening of villi in the small intestines, which affects the absorption of nutrients. A microscopic view shows the blunted villi of a celiac patient. (Credit: Wikipedia)
The biomarkers refer to a gene complex known as human leukocyte antigen (HLA) DQ2 and DQ8. Almost 100 percent of celiac patients have either of these genes. People who do not have HLA-DQ2 or DQ8 will likely never develop celiac disease.
But the genes alone don’t cause celiac disease. It is estimated that as many as a third of the people in the United States carry HLA-DQ2 or DQ8 genes but never actually develop the disease. Instead, it is thought that some additional factor is required to trigger the disease in genetically susceptible patients.
A promising study from the University of Chicago and the University of Pittsburgh School of Medicine published in Science earlier this year showed that a reaction to a common and usually harmless reovirus called TL1 may be a trigger for celiac disease, but as of now, more research needs to be done, and there is no known way to prevent or cure it.
The only current treatment is to completely avoid gluten, meaning patients with celiac disease are on a special diet for the rest of their lives. Eating at restaurants, where gluten contamination is common, is a challenge. Eating at a friend’s or relative’s house can also be a challenge because most people don’t realize how common gluten is as an additive. Ingesting even a small amount from contamination can trigger a reaction.
And although there are now substitutions, even a product labeled “gluten-free” is only required to have less than 20 parts per million of gluten (which is thought be below the threshold of triggering the disease). And gluten is even in products most people wouldn’t expect, such as medications, salad dressing, vitamins and even shampoo. The prevalence of gluten is why the idea of an enzyme that can digest it is so promising.
“It’s a shock for patients when they come to clinic and we tell them ‘you have to have to avoid gluten.’ It’s like you are handing them a harsh sentence for life,” said Tejaswi. “If you compare it to another food-related disease, like diabetes, there are so many medications for a diabetic, but there aren’t any for celiac patients. It would be the equivalent if you told a diabetic, ‘you can’t have sugar for the rest of your life.’”
Tejaswi sees tremendous potential for a therapeutic enzyme. “It would be a miracle drug if it were to happen,” he said. “For me it is something we badly need as clinicians taking care of patients. It would be very useful for patients to improve their overall quality of life. Even when patients are being careful, gluten intake does happen — it’s everywhere — and a drug like this might actually help with that.”
Tejaswi is careful to point out that a digestive enzyme for gluten wouldn’t mean patients could have carte blanche when it comes to gluten.
“I wouldn’t want patients to misunderstand a potential new drug as something they can take and eat as much pizza as they want, but it’s something that will help both physicians and the patients,” he said.
He also sees the potential for long-term improvements. “Over a long period of time this could potentially lead to fewer complications of celiac — like less osteoporosis. But for now, I think it would be a godsend to have some kind of medication for patients with celiac disease,” said Tejaswi.
From prototype to promising therapeutic drug
Although the work done for the iGEM contest was significant, it was actually just the start. The enzyme was still a prototype, nowhere near ready to be a commercial product.
“Ingrid had always been very entrepreneurial,” said Siegel. “I was going to go the professor route, and she wanted to drive the company forward.” Before Siegel left Washington to become a faculty member at UC Davis, PvP Biologics was formed as an LLC.

Ingrid Swanson Pultz
“Most of these undergraduate student projects that get to the prototype stage are then left to languish on the shelves because the students move on to their next big thing. It’s difficult to see a project with so much promise stall because there is no one to push the work forward,” said Pultz. “I sat down with Justin before he left and we made a plan for how to pursue the development of this enzyme in the short term through academia, and then in the long term with PvP Biologics.”
Pultz worked on the KumaMax prototype as a postdoc in David Baker’s lab at the University of Washington. She became a faculty member at the Institute for Protein Design — also at the University of Washington — and continued to develop the enzyme to bring it to a proof-of-concept phase.
Credit for creating the enzyme is shared—the iGEM team members, Siegel, Pultz and a few others are listed as inventors on the patents — but to be able to use and continue to develop the enzyme, PvP Biologics had to license the enzyme from the University of Washington, which owns the intellectual property. (Most of the patents are owned by the University of Washington plus one owned jointly by University of Washington and UC Davis.)
When the enzyme was ready, Pultz and Siegel started looking for venture capital funding. Siegel describes how this was right around the same time that Alvine Pharmaceuticals was finishing up clinical trials for the promising combination enzyme therapeutic from Stanford, ALV003, which contained SC PEP and EP-B2. Initially there was a lot of interest from venture capitalists. But then ALV003 had unexpectedly disappointing results with a phase two clinical trial.
“Suddenly it was really hard to get anyone interested. The venture capitalists just all sort of pulled back,” said Siegel.
Tadataka Yamada, chair of the Institute for Protein Design advisory board, stepped in to help. Yamada was a venture partner at Frazier Healthcare Partners, and had previously been the executive vice-president, chief medical and scientific officer and a board member of Takeda Pharmaceuticals. He had also served as president of the Bill and Melinda Gates Foundation Global Health Program.
Yamada helped Pultz and Siegel recruit a management team with a successful track record in commercializing pharmaceutical products. And rather than going the venture capital route, Yamada helped PvP Biologics connect directly with several pharmaceutical companies.
The strategy was a success. In January 2017, PvP Biologics announced a deal with Takeda Pharmaceuticals, which invested $35 million to move the enzyme through phase one trials with an exclusive option for purchase.
PvP is currently conducting safety testing with animals. The next step for getting Food and Drug Administration approval of the enzyme is human clinical trials, which will test for safety, efficacy and any possible side effects. The trials are scheduled to start this year.
Siegel notes that with many human drug trials the worry is about safety. A drug maker might know that a drug is effective but not know whether it is safe due to unexpected side effects. Because KumaMax is, at its most basic level, a protein that enters the stomach, and the human body is very adept at dealing with orally delivered proteins, there is less worry about whether it will be safe. “Ours is an inverse risk profile,” said Siegel. “The efficacy will be the big question.”
If effective, it could make a tremendous difference in the lives and long-term health of celiac patients.
It could also result in an extremely successful business venture. In the United States alone, there are roughly 2.5 million potential patients, which brings the possibility of KumaMax being a billion-dollar drug — but that’s only if it succeeds where other oral enzyme therapeutics to degrade gluten like ALV003 have failed. And even though KumaMax was shown to be more effective than ALV003 as far back as the iGEM contest, what works wonders in a test tube and a lab animal may simply fizzle in the human digestive system.
What’s clear is that Siegel finds the research and its potential impact very exciting.
“The real possibility to affect millions of people around the world has really driven my research in new directions,” said Siegel. “We are just starting to understand the complex relationship between our health and the food we eat. I think this will be a major step forward in developing a new paradigm for how we interact with our food.”
Resources
• UC Davis Innovation Access
• The Siegel Lab at UC Davis
• Sooraj Tejaswi, M.D., MSPH,
• PvP Biologics
Contacts
• Adam Simpson, President & Chief Executive Officer, PvP Biologics ([email protected])
• Justin Siegel, Assistant Professor, UC Davis, 530-752-9910 ([email protected])
• Lisa Howard, UC Davis Office of Research, 530-752-8117 ([email protected])
Latest News & Events